
Homebrew and Anaconda: Configuring for Apple M1 and Intel Silicon

Homebrew and Anaconda: Configuring for Apple M1 and Intel Silicon
This is a quick article that will serve as a brain dump for how I've recently configured my /bin/bash profile for using Homebrew Package Manager with both M1 and Intel Silicon (with x86 emulated using Rosetta).
Installation Locations
Depending on which CPU architecture you are installing Homebrew Package Manager with will determine where brew is installed. By default, brew will be installed to the following locations.
M1 Silicon Installation Path
If you are using brew on Apple M1 silicon, then the default installation script will install Homebrew Package Manager to the following paths.
#!/bin/bash
/opt/homebrew/bin
/opt/homebrew/CaskroomIntel Silicon Installation Path
If you are using Homebrew Package Manager on Intel silicon, or through Rosetta emulation, then you will notice that Homebrew is installed to the following paths when you use the default installation script that is available from the official homepage.
#!/bin/bash
/usr/local/bin/brew
/usr/local/CaskroomConfiguring Bash
Now that we know where brew is installed, we can configure bash to automatically load the relevant shell environment depending on which architecture is being used. This is useful if you have multiple copies of iTerm2 (for example), whereby one copy may be configured to launch with Rosetta emulation. If your Terminal instance (including iTerm2) is launched with Rosetta emulation, it will automatically attempt to use Homebrew at the listed installation path above (under "Intel Silicon"). However, because both installations are at different paths, we need some logic to detect which architecture (emulated or not) is being actively used, so we know which shell environment to load.
Refer to the logic below. It uses a command called "uname" to detect which CPU architecture is actively in use. Based on the resulting value, it will then load the relevant shell environment. x86_64 is considered to be Intel Silicon, and arm64 is considered to be Apple M1.
Homebrew Shell Environment
The snippet below describes the logic for automatically loading Homebrew shell environment depending on the architecture.
#!/bin/bash
if [[ "$(uname -m)" == "x86_64" ]]; then
echo "Loading: Homebrew (x86)"
eval "$(/usr/local/bin/brew shellenv)"
CONDA_BREW_PATH=/usr/local/Caskroom/miniconda
else
echo "Loading: Homebrew (ARM)"
eval "$(/opt/homebrew/bin/brew shellenv)"
CONDA_BREW_PATH=/opt/homebrew/Caskroom/miniconda
fi
Anaconda Shell Environment
The snippet below describes the logic (typically generated by Anaconda) for loading the relevant shell environment.
#!/bin/bash
if [[ "$(uname -m)" == "x86_64" ]]; then
# >>> conda initialize >>>
# !! Contents within this block are managed by 'conda init' !!
__conda_setup="$('/usr/local/Caskroom/miniforge/base/bin/conda' 'shell.bash' 'hook' 2> /dev/null)"
if [ $? -eq 0 ]; then
eval "$__conda_setup"
else
if [ -f "/usr/local/Caskroom/miniforge/base/etc/profile.d/conda.sh" ]; then
. "/usr/local/Caskroom/miniforge/base/etc/profile.d/conda.sh" # commented out by conda initialize
else
export PATH="/usr/local/Caskroom/miniforge/base/bin:$PATH" # commented out by conda initialize
fi
fi
unset __conda_setup
# <<< conda initialize <<<
elif [[ "$(uname -m)" == "arm64" ]]; then
# >>> conda initialize >>>
# !! Contents within this block are managed by 'conda init' !!
__conda_setup="$('/opt/homebrew/Caskroom/miniforge/base/bin/conda' 'shell.bash' 'hook' 2> /dev/null)"
if [ $? -eq 0 ]; then
eval "$__conda_setup"
else
if [ -f "/opt/homebrew/Caskroom/miniforge/base/etc/profile.d/conda.sh" ]; then
. "/opt/homebrew/Caskroom/miniforge/base/etc/profile.d/conda.sh" # commented out by conda initialize
else
export PATH="/opt/homebrew/Caskroom/miniforge/base/bin:$PATH" # commented out by conda initialize
fi
fi
unset __conda_setup
# <<< conda initialize <<<
fiConfiguring iTerm to Launch with Rosetta Emulation
If you are running on Apple M1 silicon, you can still run the same applications with x86 emulation by using Rosetta. You can install Rosetta from the command-line by running the following command.
#!/bin/bash
/usr/sbin/softwareupdate --install-rosetta --agree-to-licenseAnd now you can create a copy of your iTerm installation, and modify the copy to launch with Rosetta emulation. Refer to the screenshots below for greater reference.
A picture of "Applications" in macOS finder, with a duplicate or copy of iTerm.
With your duplicate instance of iTerm, you can then modify the properties so that it launches with Rosetta emulation.
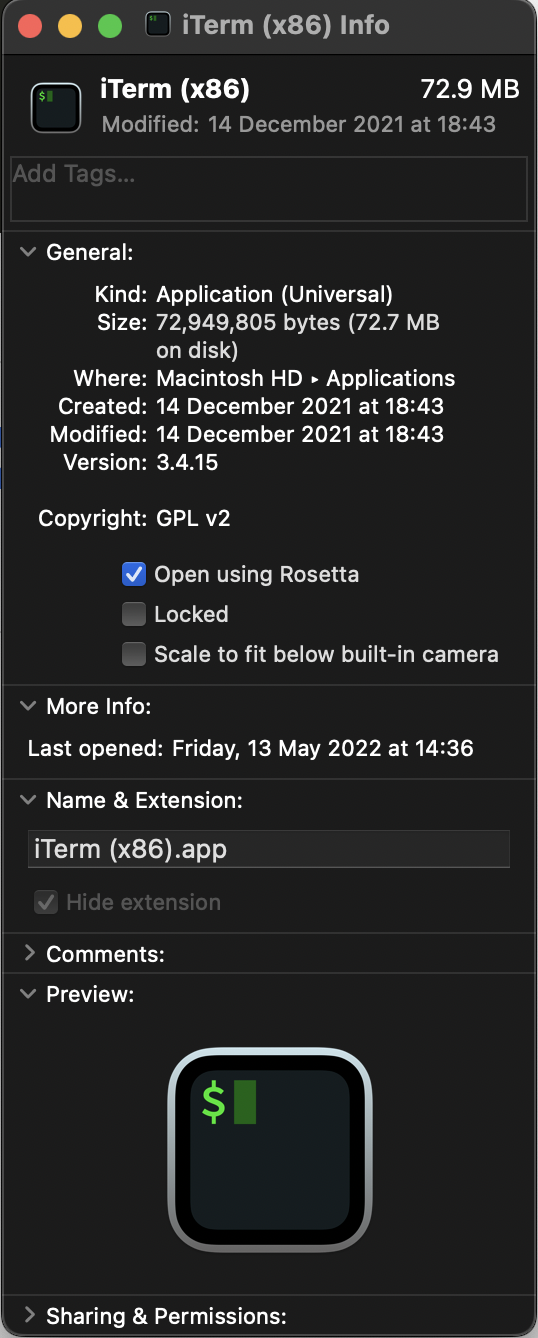
After you've duplicated your iTerm installation and renamed it appropriately, you will then have to modify the properties so that it launches with Rosetta.
Additionally, if you want to be able to interchangeably use either Homebrew installations while using one kind of architecture over the other, then you can make use of the following aliases that can be added to your ~/.profile file. It should be noted that this snippet includes support for pyenv, a tool that is used for managing multiple installations of Python on the same machine.
#!/bin/bash
eval "$(pyenv init -)"
eval "$(pyenv virtualenv-init -)"
alias ibrew='arch -x86_64 /usr/local/bin/brew'
alias mbrew='arch -arm64e /opt/homebrew/bin/brew'There you have it. You should be able to use Homebrew with both architectures from the same Apple M1 machine.
Comments
Comments
If you call any of us in Hawaii, we will be glad to tell you about our state laws, so that you can better prepare your patients <a href=http://cialiss.hair>cialis buy online usa</a>
Howdy I am so glad I found your blog, I really found you by accident, while I was
browsing on Askjeeve for something else, Nonetheless I am
here now and would just like to say thank you for a
tremendous post and a all round thrilling blog (I also love the theme/design),
I don’t have time to read through it all at the moment but I have bookmarked it and
also added your RSS feeds, so when I have time I will
be back to read a lot more, Please do keep up the fantastic b.
Great post. I was checking constantly this blog and I am impressed!
Very useful info specially the final section :) I handle such information a
lot. I was looking for this certain info
for a long time. Thanks and good luck.
Hello there! <a href=http://edpill.online/>ed pills online</a> very good web site http://edpill.online
Howdy! <a href=http://edpill.online/>ed pills</a> great website http://edpill.online
Howdy! <a href=http://edpill.online/>treatment for erectile dysfunction</a> very good internet site http://edpill.online
Peripheral cannabinoid receptor, CB2, regulates bone mass <a href=https://acialis.mom>cialis buy</a>
<a href=http://ciali.sbs>cialis cheapest online prices</a> This medication makes me extremely tired and left without energy
Ηi! This is my fiгst comment here sօ I just wanted to give
a quick shout out and say I genuinely enjoy rеading through yoᥙr blog posts.
Can yоᥙ suggest any other blogs/websites/fօrums that deal
with the same subjects? Thank you!
thank you very much
_________________
<a href=http://ipl.kzkkslots4.fun/ipl_app_key_eng.html>ipl live match video app free</a>